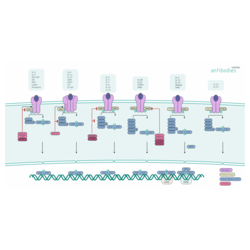
JAK-STAT Signaling
JAK-STAT signaling in vertebrates relies on a network of protein kinases and transcription factors to integrate signals from various receptor systems. The multitude of stimuli include cytokines, growth factors, and hormones, binding of which ultimately effect processes such as immune response regulation and cell growth, survival, and differentiation.
The highly conserved pathway involves essentially three levels of processing of the incoming information depending on the function of the respective components:
Binding of the ligand to the receptor triggers conformational changes of the receptor molecule(s). This steric change of the receptor brings two Janus Kinases (JAK) bound to the receptor or receptor subunits into close proximity, thus enabling trans-phosphorylation. The activated JAKs phosphorylate subsequently additional targets. The major phosphorylation targets are Signal transducer and activator of transcription (STAT). These transcription factors are inactive in the cytoplasm until phosphorylation by JAKs. Once the conserved tyrosine toward the C-terminus of the STAT has been phosphorylated, it can act as dimerization interface in conjunction with SH2 domains of another STAT. These activated STAT dimers are then translocated to the nucleus and bind to specific DNA motifs to activate target gene transcription.
Besides, negative regulation of these processes takes place on multiple levels:
- Suppressors of cytokine signaling (SOCS) gene transcription is stimulated by activated STATs. SOCS inactivate signaling through binding to the phosphorylated JAKs or receptors or facilitate JAK ubiquitination.
- Protein inhibitor of activated STATs (PIAS) bind to activated STATs and prevent them from binding DNA.
- Protein tyrosine phosphatases (PTP) reverse the activity of JAKs.
The JAK-STAT signaling pathway transduces downstream of multiple cytokines critical to the pathogenesis of immune-mediated disease. It has been suggested that patients with mutations in STAT1 and STAT2 are often more likely to develop infections from bacteria and viruses. Since many cytokines function through the STAT3 transcription factor, STAT3 plays a significant role in maintaining skin immunity. In addition, because patients with JAK3 gene mutations have no functional T cells, B cells or NK cells, they would more likely to develop skin infections. Mutations of the STAT5 protein, which can signal with JAK3, has been shown to result in autoimmune disorders. Also, STAT4 mutations have been associated with rheumatoid arthritis, and STAT6 mutations are linked to asthma.
Current research on this pathway has been focusing on the inflammatory and neoplastic diseases and related drugs. One example of a JAK inhibitor drug is Ruxolitinib, which is used as a JAK2 inhibitor. JAK1 and JAK3 inhibitor Tofacitinib has been used for psoriasis and rheumatoid arthritis treatment and is under investigation for many other immune-mediated diseases including systemic lupus erythematosus, inflammatory bowel disease, and rare autoinflammatory diseases with a type 1 interferon signature. It has been reported that therapies which target STAT3 can improve the survival of patients with cancer.
In the course of the COVID-19 pandemic several JAK inhibtors have been approved for treatment of COVID-19 induced cytokine storm, an excessive inflammatory reaction in which cytokines are rapidly produced in large amount in response to an infection.
The prototypical JAK-STAT signaling pathway is rather linear. There is however considerable crosstalk with other signaling cascades like MAPK pathways, PI3K signaling and JAK independent STAT phosphorylation through receptor tyrosine kinase (RTKs).
References:
- : "JAK-STAT signaling: from interferons to cytokines." in: The Journal of biological chemistry, Vol. 282, Issue 28, pp. 20059-63, (2007) (PubMed).
- : "Biology and significance of the JAK/STAT signalling pathways." in: Growth factors (Chur, Switzerland), Vol. 30, Issue 2, pp. 88-106, (2012) (PubMed).
- : "Mechanisms of Jak/STAT signaling in immunity and disease." in: Journal of immunology (Baltimore, Md. : 1950), Vol. 194, Issue 1, pp. 21-7, (2016) (PubMed).
- : "The JAK-STAT signaling pathway: input and output integration." in: Journal of immunology (Baltimore, Md. : 1950), Vol. 178, Issue 5, pp. 2623-9, (2007) (PubMed).
- : "Series introduction. JAK-STAT signaling in human disease." in: The Journal of clinical investigation, Vol. 109, Issue 9, pp. 1133-7, (2002) (PubMed).
- : "Protein tyrosine phosphatases in the JAK/STAT pathway." in: Frontiers in bioscience : a journal and virtual library, Vol. 13, pp. 4925-32, (2008) (PubMed).
- : "JAK-STAT Signaling as a Target for Inflammatory and Autoimmune Diseases: Current and Future Prospects." in: Drugs, Vol. 77, Issue 5, pp. 521-546, (2017) (PubMed).
- : "Loss of function mutations in PTPN6 promote STAT3 deregulation via JAK3 kinase in diffuse large B-cell lymphoma." in: Oncotarget, Vol. 6, Issue 42, pp. 44703-13, (2016) (PubMed).
- : "Targeting JAK-STAT Signaling to Control Cytokine Release Syndrome in COVID-19." in: Trends in pharmacological sciences, Vol. 41, Issue 8, pp. 531-543, (2020) (PubMed).
- : "SARS-CoV-2 drives JAK1/2-dependent local complement hyperactivation." in: Science immunology, Vol. 6, Issue 58, (2021) (PubMed).
References:
- (11)A. Kukowka et al. in: "The role of janus kinases in the treatment of autoimmune skin diseases". Farmacja Polska (2021).