Production of functional proteins is a crucial step for scientist in order to investigate the function and structure requires of proteins of interest. Moreover, recombinant enzymes are used in the food and chemical industries, and high-quality proteins are required for diagnostic, therapeutic, and pharmaceutical applications. Chemical synthesis is not advanced enough to correctly synthesize complex protein structures. Instead living organisms are manipulated to build and construct proteins based on supplied genetic information in form of DNA. The structure of DNA is simple enough to engineer DNA templates with desired coded functions like add-on reporter or affinity tag sequences. Proteins generated from artificial templates are called recombinant proteins. This article highlights advantages and disadvantages of microbial and mammalian cell-based as well as cell-free plant-based recombinant protein expression sytems.
Recombinant Protein Expression
The first established protocol for recombinant protein expression utilizes transfection of host cells with a DNA vector that contains the template and their subsequent proliferation. The transcription and translation machinery in the host cells produce multiple copies of the protein of interest which can be accessed via lyse and subsequent purification.
Since then a variety of expression systems and protocols emerged, prokaryotic as well as eukaryotic in vivo protein expression systems are widely used. Protein type, requirements for functional activity and the desired yield influence the choice of the system. Bacterial, yeast, insect, mammalian or plant - each system has advantages and challenges, and choosing the right system for the specific application is important for successful recombinant protein expression. This following table provides an overview about advantages and challenges in recombinant protein expression systems.
Comparison of Protein Expression Systems
| Properties | E. coli | Yeast | Insect Cells | Mammalian Cells | Plant Cells |
| Cell Growth | fast (30 min) | fast (90 min) | slow (18-24 h) | slow (24 h) | cell-free |
| Culture Conditions | simple | simple | complex | complex | simple |
| Expression Level | high | average | average | low | high |
| Extracellular Expression | secretion to periplasm | secretion to medium | secretion to medium | secretion to medium | cell-free |
Post-translational Modification
After translation as a last step of maturation, proteins are modified in various ways to complete their structure, designate their location or regulate their activity within the cell. Post-translational modifications (PTMs) are various additions or alterations to the chemical structure and are critical features of the overall cell biology. These modifications can include removal of segments using peptidases, addition of phosphate, sugar or lipids to specific amino acids and glycosylation. PTMs can occur on the amino acid side chains or at the protein's C- or N- termini.
| Posttranslational Modifications | E. coli | Yeast | Insect Cells | Mammalian Cells | Plant Cells |
| Protein Folding | refolding usually required | refolding may be required | proper folding | proper folding | proper folding |
| N-linked Glycosylation | none | high mannose | simple, no sialic acid | complex | complex |
| O-linked Glycosylation | no | yes | yes | yes | yes |
| Phosphorylation | no | yes | yes | yes | yes |
| Acetylation | no | yes | yes | yes | yes |
| Acylation | no | yes | yes | yes | yes |
| Gamma-Carboxylation | no | no | no | yes | yes |
After the protein is synthesized disulfide bonds are formed and the protein folds into its three dimensional state. The simplest of these modifications is the removal of the N-terminal methionine residue, which can occur in all organisms. Methionine excision is regulated at various levels and plays an important role in controlling protein turnover, the balance between protein synthesis and protein degradation.
Signal peptides are 20 to 100 amino acids long sequences majorly N-terminal attached to a protein. The properties of the amino acids are the significant factors determining interaction with the protein transport system, hence the destination to which that protein is delivered. Proteins without remain in the cytoplasm. Different signal sequences can be combined and allow dual targeting of proteins into two compartments at a time. Signal sequences and transport process differ between kingdoms. This represents a challenge in recombinant expression of functional proteins.
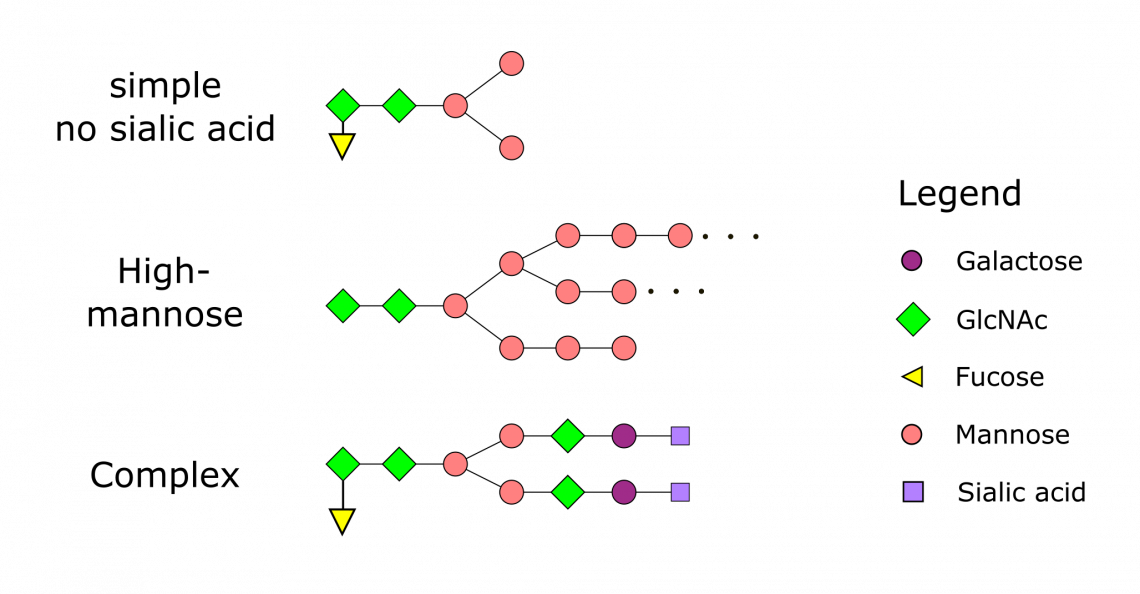
Fig. 1: N-Glycosylation Patterns
Glycosylation
Similar to this, more complex modifications, like N- and O-glycosylation, phosphorylation, are exclusively carried out by eukaryotic cells. Together with nitrosylation, GTP binding and acetylation of histones function and expression of the protein are regulated. Prenylation supports membrane localization of proteins.
The nature of N-linked glycans attached to a glycoprotein is determined by the protein and the cell in which it is expressed. Eukaryotic expression hosts such as yeast and animal cells, have different glycosylation patterns. While yeast glycosylation usually shows a high proportion of mannose, insect cells pattern is rather simple and without sialic acid. Animals and human are more alike, however they show a lot of variance, the patterns are more complicated with galactose, sialic acid and fucose build in. The patterns also vary across species. Different species synthesize different types of N-linked glycan.
Glycosylation patterns are involved in cell migration and are important for both the structure and function of the protein. They can promote protein folding and improve stability. Immunoglobulins like IgE, IgM or IgG utiize glycosylation – a change in affinities for Fc and other immune receptors generate unique effector functions. In the evolving market of recombinant therapeutic monoclonal antibodies like
and . These slight changes are of immense importance as they can trigger potential immunogenic reactions. Chinese hamster ovary (CHO) cells have established themselves as go-to cell line for biological and medical research and commercially in the production of therapeutic proteins. They show high agreement in pattern with human cells. However, glycans produced in these systems can differ from glycans produced in humans, as they can be capped with both N-glycolylneuraminic acid (Neu5Gc) and N-acetylneuraminic acid (Neu5Ac), whereas human cells only produce glycoproteins containing N-acetylneuraminic acid. Additionally galactose-alpha-1,3-galactose epitopes from glycoproteins produced in animal cells can induce severe anaphylactic shocks. Scientist try to optimize the systems by eliminating the pathways that produce these glycan structures through genetic knockouts or overexpression.Check table 2 to find out which expression system carries out the PTMs you are looking for.
Bacterial protein Expression
Bacterial protein expression systems are well-utilized in recombinant protein experiments especially on a bigger scale. Simple culture conditions the fast and simple growth of bacteria lead to high scalability together with low production costs.
The system offers sufficient quality for most applications like antibody generation, functional assays and protein interaction studies. However, eukaryotic proteins expressed in bacteria often are non-functional because the cells are not equipped to accomplish the required PTMs or folding steps. Quality concerns in bacterial protein expression typically occur with large proteins (> 50 kDa), complex protein structures like multiple domains and transmembran spans which require multiple signal peptides and PTMs like disulfide bonds or glycosylation.
Another obstacle is the aggregation of recombinant disulfide-bonded proteins in E. coli in so called inclusion bodies. E. coli’s cytoplasm is maintained in a reduced state that strongly disfavors the formation of stable disulfide bonds in proteins.
The aggregation is a direct consequence of the lack of disulfide bond formation, 70-80% of recombinant proteins expressed in E. coli are contained in inclusion bodies. Purification is time consuming and usually requires two main steps: extraction of inclusion bodies from the bacteria followed by the solubilisation of the purified inclusion bodies.
Insect Protein Expression
To generate large amounts of a functional recombinant protein, many investigators have turned to baculovirus expression systems. In comparison to bacterial systems, insect protein expression shows closer relation to the mammalian processes. This is why these systems are particularly well-suited for proteins that are difficult to express in bacteria due to size, complexity, or posttranslational processing requirements. These systems can be easily scaled up and adapted to high-density suspension culture for large-scale
Among the eukaryotic expression systems, the baculovirus-infected insect cell platform has gained particular attention, resulting in the development and implementation of multiple strategies for protein expression. It takes advantage of the ability of baculovirus to insert genes into its target cell and induce protein expression this gene. The most common species of baculoviruses used for protein expression studies is the
.Numerous insect cells have been developed into cell lines, such as fruit flies, mosquitoes, and silkworms. Over time tissue of the cabbage looper (Trichoplusia ni) has established itself as cell line of choice. It is particularly useful for its fast growth rate and less reliance upon insect haemolymph in the medium. The Trichoplusia ni cell line High Five (officially called BTI-Tn-5B1-4) expresses consistently 5 to 10 times more recombinant protein than other lepidopteran cell lines, such as Sf9 cells. They can be grown in the absence of serum which lowers the production costs by a big margin, and can be cultured in a loose attached state or in suspension.
Two approaches were developed for expression in insect cells, secreted and extracellular. The choice of targeting of a given protein for expression is determined by its cellular localization in the native state. Proteins that function extracellularly are expressed as secreted proteins and are collected from the medium. For secreted expression a signal peptide has to be genetically engineered to the insert of (His) tag, TEV protease recognition site and desired gene. On the other hand, proteins that function in the nucleus or in the cytoplasm are expressed intracellularly.
Downfalls of recombinant protein production in insect expression systems are long production time and relative high costs. Compare advantages and disadvantages of protein expression systems at a glance, or browse our broad offer of recombinant proteins with unmatched quality expressed in insect cells - available for purchase at antibodies-online
Mammalian Protein Expression
Even though many mammalian proteins have been successfully produced in bacteria, yeast, or insect cells, the demand for mammalian cell cultures is rising. Proteins in appropriate quantity and quality are an essential requirement of the present time. Expression systems utilizing mammalian cells for recombinant proteins are able to introduce proper protein folding, post-translational modifications, and product assembly, which are important for complete functional activity. However, these benefits are coupled with more demanding culture conditions and high production costs.
Protein expression in mammalian cells can be achieved via plasmid transient transfection, cell line establishment, retrovirus or lentivirus transduction or modified mRNA transfection.
Mammalian expression systems are able to introduce either transiently or stable cell lines, where the expression construct is integrated into the host genome. While stable cell lines can be used over several experiments, transient production can generate large amounts of protein in one to two weeks. They utilize suspension cultures and can produce gram-per-liter yields in contrast normal mammalian cell lines which usually have slow cell growth and low expression levels.
Plant Protein Expression
The plant transient expression system has emerged as a promising platform for producing a diverse range of vaccines, therapeutic proteins, enzymes, and antibodies. Plant-based expression systems offer several potential advantages, including safety, and cost-effectiveness in generating biologically active products. Moreover, these systems provide eukaryotic PTMs, enhancing the functionality of expressed proteins. This is particularly important in the context therapeutic proteins and human enzymes.
Cell-free protein expression (CFPE) is a method of protein production via utilization of the RNA and protein synthesis machinery of cells in a system without cellular membranes and a functional genome. This in vitro approach enables the flexible expression of a diverse array of proteins under varying conditions. An intriguing feature of cell-free protein expression lies in its ability to facilitate the production of proteins that are challenging or infeasible to express within living cells, especially those that induce toxic effects. Despite their limited compatibility with cell-based expression methods, these proteins frequently serve as drug targets or hold potential as therapeutic agents.
ALiCE® (Almost Living Cell-Free Expression) is an eukaryotic protein expression platform able to produce proteins at scale. The system combines advantages of Nicotiana. Tabacum as plant expression system with the rapid expression of CFPE. ALiCE® has a high protein yield averaging about 2 grams per liter within a very short time frame of 48 hours. For industrial purposes the yield can be upscaled up to 10 liter. Addtionally, ALiCE® is able to attach eukaryotic PTMs like complex glycolisation patterns to the proteins.
References
- : "Selecting an appropriate method for expressing a recombinant protein." in: Methods in enzymology, Vol. 463, pp. 131-47, (2010) (PubMed).
- : "Recombinant protein expression in Escherichia coli." in: Current opinion in biotechnology, Vol. 10, Issue 5, pp. 411-21, (1999) (PubMed).
- : "Baculovirus as versatile vectors for protein expression in insect and mammalian cells." in: Nature biotechnology, Vol. 23, Issue 5, pp. 567-75, (2005) (PubMed).
- : "Mammalian cell protein expression for biopharmaceutical production." in: Biotechnology advances, Vol. 30, Issue 5, pp. 1158-70, (2012) (PubMed).
- : "Overview of protein expression by mammalian cells." in: Current protocols in protein science, Vol. Chapter 5, Issue 1, pp. Unit5.9, (2008) (PubMed).
- : "Expression of mammalian proteins for diagnostics and therapeutics: a review." in: Molecular biology reports, Vol. 49, Issue 11, pp. 10593-10608, (2022) (PubMed).
- : "Improvement of the transient expression system for production of recombinant proteins in plants." in: Scientific reports, Vol. 8, Issue 1, pp. 4755, (2019) (PubMed).
- : "Post-translational modification of plant-made foreign proteins; glycosylation and beyond." in: Biotechnology advances, Vol. 30, Issue 2, pp. 410-8, (2012) (PubMed).

Creative mind of antibodies-online with a keen eye for details. Proficient in the field of life-science with a passion for plant biotechnology and clinical study design. Responsible for illustrated and written content at antibodies-online as well as supervision of the antibodies-online scholarship program.
Go to author page



