SARS-CoV-2 Neutralizing Antibodies
Neutralizing antibodies are of particular interest to scientists. They efficiently stop the infection by blocking the interaction between the SARS-CoV-2 virus and the host cells. Most neutralizing antibodies respond to the receptor binding domain (RBD) of the spike protein, which binds directly to the cell surface receptor ACE2. antibodies-online.com offers two nAbs based on clone CR3022 as well as clones based on MM43 and MM117
See all Neutralizing SARS-CoV-2 Antibodies
One of the looming questions regarding newly emerging variants of SARS-CoV-2 is, whether existing antibody based assays and pharmaceuticals remain useable. In collaboration with Nanotemper we measured the affinity of our S protein RBD antibody CR3022 (ABIN6952546) to the canonical trimeric SARS-CoV-2 S protein and those of the variants of concern B.1.1.7 / alpha, B.1.351 / beta, B.1.617.2 / delta, B.1.617.1 / kappa and P.1 / gamma by microscale thermophoresis (MST). In a second iteration we acknowledged the dominant B.1.1.529 / omicron variant and extended the antibody selection with neutralizing antibodies MM43 (ABIN7036075) and MM117 . Discover the results down below, either summarized in the table or in extensive form with attached MST binding curves. Our webinar provides more background information about the experiments and impact on assays and vaccine production. We aim to keep the comparison measurements up to date in respect to new antibodies and emerging variants.
MST analysis of SARS-CoV-2 nAbs
Webinar: "Effect of SARS-CoV-2 Variants for Assays and Vaccines"
SARS-CoV-2 Neutralizing Antibodies and corresponding Binding Preferences
| Canonical | Delta B.1.617.2 | Omicron B.1.1.529 | |
|---|---|---|---|
| CR3022 ABIN6952546 |
++ | + | + |
| MM43 |
+++ | ++ | - |
| MM117 | + | + | ++ |
Microscale Thermophoresis analysis of SARS-CoV-2 nAbs
CR3022
The antibody clone CR3022 (ABIN6952546), which is the basis for both antibodies, was originally isolated from a convalescent SARS patient from Singapore1. The clone was demonstrated to be effective in neutralization assays for different SARS-CoV strains in synergy with other RBD-targeting antibodies. Its epitope does not overlap with the angiotensin-converting enzyme 2 (ACE2) binding site, thus leaving it accessible for other neutralizing antibodies. Since the outbreak of COVID-19, CR3022 has been demonstrated to bind the SARS-CoV-2 S protein RBD in a similar fashion2. Crystallization assays of CR3022 bound to its SARS-CoV-2 target have provided important insights into possible attack points for therapeutics against this virus3. Moreover, CR3022 has been used as a positive control in serological assays to detect antibodies in human serum that bind SARS-CoV-2 S-protein4.
A higher dissociation constant is indicative of a weaker binding, i.e. CR3022 binding of the B.1.351 S protein is significantly weaker than of the other three variants. However, a KD in the nanomolar range does indicate a high affinity of the antibody to all four tested S proteins. Accordingly, the neutralizing antibody remains an important tool for SARS-CoV-2 immunoassays.
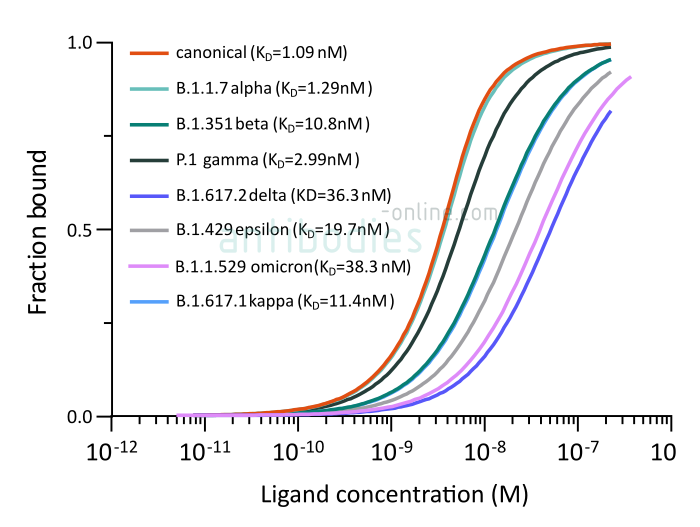
Fig. 1: Microscale thermophoresis measurements of binding of SARS-CoV-2 Spike S1 antibody (RBD) CR3022 (ABIN6952546) to SARS-CoV-2 Spike (Trimer) from wt SARS-CoV-2 (ABIN6952670) or variants B.1.1.7 alpha (ABIN6963742), B.1.351 beta (ABIN6963740), P.1 gamma (ABIN6964443), B.1.6172 delta (ABIN6999328), B.1.429 epsilon (ABIN6972926), B.1.617.1 kappa (ABIN6976302) and B.1.1.529 omicron (ABIN7041433). The determined dissociation constants KD are indicated.
MM117
MM117 is a monoclonal antibody raised in mice that binds the SARS-CoV-2 B.1.1.529 omicron S protein more tightly than that of other variants.
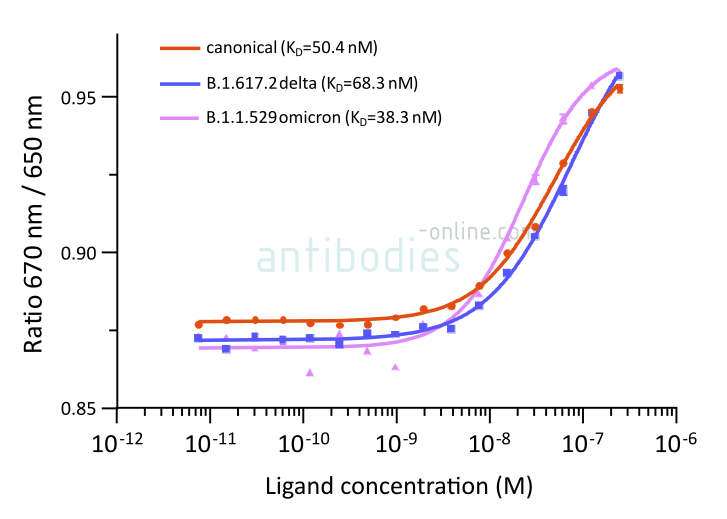
Fig. 2: Microscale thermophoresis measurements of binding of SARS-CoV-2 Spike S1 antibody (RBD) MM117 to SARS-CoV-2 Spike (Trimer) proteins of various lineages, including B.1.617.2 (delta) and B.1.1.529 (omicron). The determined dissociation constants KD are indicated.
MM43
The murine monoclonal antibody MM43 (ABIN7036075) on the other hand bind the canonical S protein more tightly than CR3022 and MM117. Binding to the SARS-CoV-2 B.1.617.2 delta S protein is also very good. However, binding of this antibody to the omicron variant’s S protein is completely abolished due to the protein’s mutations.
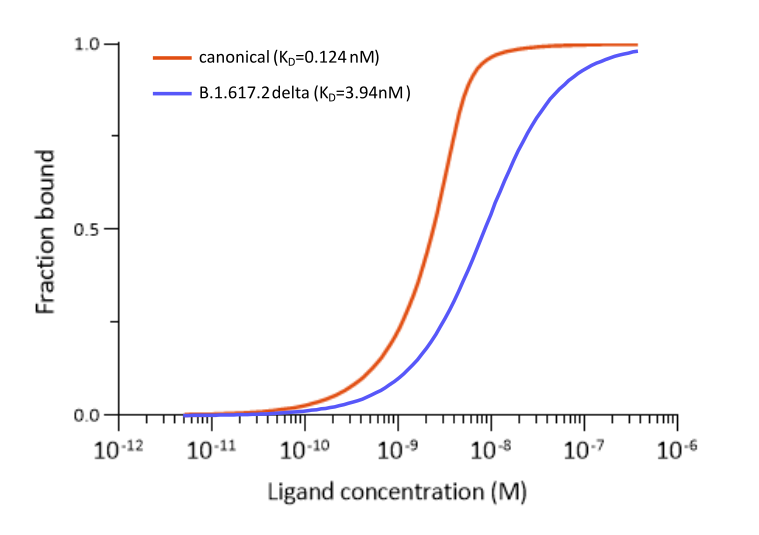
Fig. 3: Microscale thermophoresis measurements of binding of SARS-CoV-2 Spike S1 antibody (RBD) MM43 (ABIN7036075) to SARS-CoV-2 Spike (Trimer) of various lineages. Binds SARS-CoV-2 S proteins of various lineages, including B.1.617.2 (delta). Does NOT bind the S protein of SARS-CoV-2 B.1.1.529 (omicron).The determined dissociation constants KD are indicated.
Available SARS-CoV-2 Neutralizing Antibodies
- (8)
- (15)
- (1)
- (5)
- (3)
- (1)
Thought Experiment prior to MST Measurement
The SARS-CoV-2 S Protein is essential for the virions’ contact to the host cell via the ACE2 receptor. ACE2 contact points are situated between amino acids K417 and Y505 of the S Protein Receptor Binding Domain (RBD). Neutralizing antibodies (nAbs) targeting the RBD disrupt this interaction and can thus impede SARS-CoV-2 interaction. Based on the known contact points of the neutralizing antibody CR3022 with the S protein we had predicted that the antibody would still bind the protein from variants of concern (Fig.4).6,7,8,9
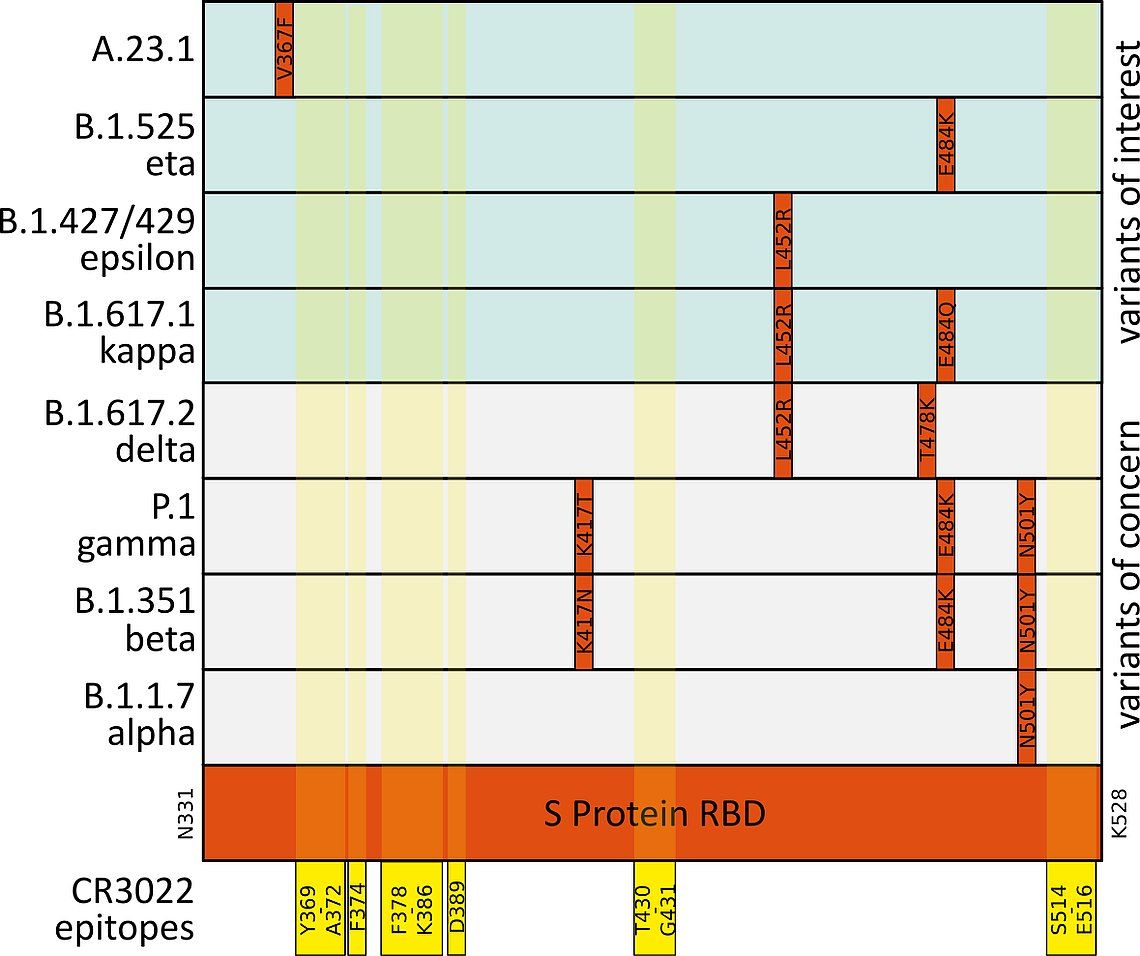
Fig. 4: Mutations (indicated in red) in the SARS-CoV-2 S Protein receptor binding domain (RBD, mint) in variants of concern (light grey) and variants of note (light mint). Epitopes of the neutralizing antibody CR3022 ABIN6952546 are highlighted in yellow.6,7,8,9
This prediction was confirmed by the MST measurements. The B.1.1.529 / omicron variant shows, in comparison, the most mutations in RBD site. Nevertheless binding of CR3022 could be confirmed experimentally. Deeper background information can be found in our webinar "Effect of Different SARS-CoV-2 Variants on COVID-19 Assays and Vaccines".
Webinar: "Effect of Different SARS-CoV-2 Variants on COVID-19 Assays and Vaccines"
References
- (1) ter Meulen, J. et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 3, e237–e237 (2006).
- (2) Tian, X. et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 9, 382–385 (2020).
- (3) Yuan, M. et al. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science 80. eabb7269 . doi:10.1126/science.abb7269 (2020).
- (4) Stadlbauer, D. et al. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 57, e100 (2020).
- (5) Thao, T. et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform Nature. (2020).
- (6) Barnes CO et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature (2020). doi:10.1038/s41586-020-2852-1
- (7) Cheng MH et al. Impact of South African 501.V2 Variant on SARS-CoV-2 Spike Infectivity and Neutralization: A Structure-based Computational Assessment. bioRxiv 2021.01.10.426143 (2021). doi:10.1101/2021.01.10.426143
- (8) Garcia-Beltran WF et al. Circulating SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. medRxiv 2021.02.14.21251704 (2021). doi:10.1101/2021.02.14.21251704
- (9) Yuan M et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633 (2020).
- (10) Taylor, P.C., Adams, A.C., Hufford, M.M. et al. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat Rev Immunol (2021). https://doi.org/10.1038/s41577-021-00542-x
- (11) Emily Engelhart, Randolph Lopez, Ryan Emerson, Charles Lin, Colleen Shikany, Daniel Guion, Mary Kelley, David Younger. Massively Multiplexed Affinity Characterization of Therapeutic Antibodies Against SARS-CoV-2 Variants. Preprint. bioRxiv 2021.04.27.440939; doi: https://doi.org/10.1101/2021.04.27.440939

Goal-oriented, time line driven scientist, proficiently trained in different academic institutions in Germany, France and the USA. Experienced in the life sciences e-commerce environment with a focus on product development and customer relation management.
Go to author page




