Histone 3 antibody (H3K27ac)
-
- Target See all Histone 3 (H3) Antibodies
- Histone 3 (H3)
-
Binding Specificity
- H3K27ac
-
Reactivity
- Human, Saccharomyces cerevisiae
-
Host
- Rabbit
-
Clonality
- Polyclonal
-
Conjugate
- This Histone 3 antibody is un-conjugated
-
Application
- Western Blotting (WB), Immunofluorescence (IF), Chromatin Immunoprecipitation (ChIP), Dot Blot (DB), Immunocytochemistry (ICC), ChIP DNA-Sequencing (ChIP-seq), Cleavage Under Targets and Release Using Nuclease (CUT&RUN), Cleavage Under Targets and Tagmentation (CUT&Tag)
- Purification
- Protein A Chromatography
- Immunogen
- This Histone H3 acetylLys27 antibody was raised against a peptide including acetyllysine 27 of histone H3.
- Isotype
- IgG
- Top Product
- Discover our top product H3 Primary Antibody
-
-
- Application Notes
-
Recommended starting concentrations are
ChIP: 10 µg per ChIP
ChIP-Seq: 5 µg each
ICC/IF: 1 - 5 µg/mL dilution
WB: 0.1 - 1 µg/mL dilution
CUT&RUN: 1:100
CUT&Tag: 1 µg/50 µL reaction
CUT&RUN: 2 µL/200 µL reaction
Optimal working dilution should be determined by the investigator. - Restrictions
- For Research Use only
-
- by
- Anna Nordin and Claudio Cantù; Cantù Lab, Gene Regulation during Development and Disease, Linköping University
- No.
- #104489
- Date
- 08/14/2023
- Antigen
- H3K27ac
- Lot Number
- 31521015
- Method validated
- Cleavage Under Targets and Release Using Nuclease
- Positive Control
Polyclonal rabbit anti-H3K4me (antibodies-online, ABIN3023251)
- Negative Control
Polyclonal guinea pig anti-rabbit IgG (antibodies-online, ABIN101961)
- Notes
Passed. ABIN2668475 allows for specific targeting of H3K27ac in human cells using CUT&RUN.
- Primary Antibody
- ABIN2668475
- Secondary Antibody
- Full Protocol
- Cell harvest
- Harvest 50,000 human fibroblast cells per antibody.
- Centrifuge cell solution 3 min at 600 x g at RT.
- Remove the liquid carefully.
- Gently resuspend cells in 1 mL of Nuclear Extraction Buffer (20 mM HEPES-KOH pH 8.2, 20% Glycerol, 0,05% IGEPAL, 0.5 mM Spermidine, 10 mM KCl, Roche Complete Protease Inhibitor EDTA-free).
- Move the solution to a 2 mL centrifuge tube.
- Pellet the nuclei 800 x g for 5 min.
- Repeat the NE Buffer wash twice for a total of three washes.
- Resuspend the nuclei in 20 µL NE Buffer per sample.
- Concanavalin A beads preparation
- Prepare one 2 mL microcentrifuge tube.
- Gently resuspend the magnetic Concanavalin A Beads (antibodies-online, ABIN6923139).
- Pipette 10 µL Con A Beads slurry for each sample into the 1.5 mL microcentrifuge tube.
- Place the tube on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tube from the magnetic stand.
- Pipette 1 mL Binding Buffer (20 mM HEPES pH 7.5, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2) into each tube and resuspend ConA beads by gentle pipetting.
- Spin down the liquid from the lid with a quick pulse in a table-top centrifuge.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tube from the magnetic stand.
- Repeat twice for a total of three washes.
- Gently resuspend the ConA Beads in a volume of Binding Buffer corresponding to the original volume of bead slurry, i.e. 10 µL per sample.
- Cell immobilization – binding to Concanavalin A beads
- Carefully vortex the nuclei suspension and add 10 µL of the Con A beads in Binding Buffer to the cell suspension for each sample.
- Close tube tightly incubates 10 min at 4 °C.
- Put the 2 mL tube on the magnet stand and when the liquid is clear remove the supernatant.
- Resuspend the beads in 1 mL of EDTA wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, Roche Complete Protease Inhibitor EDTA-free, 2mM EDTA).
- Incubate 5 min at RT.
- Place the tube on the magnet stand and when the liquid is clear remove the supernatant.
- Resuspend the beads in 200µl of Wash Buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, Roche Complete Protease Inhibitor EDTA-free) for each sample.
- Cell permeabilization and primary antibody binding
- Divide nuclei suspension into separate PCR tubes, one for each antibody (200 µL per sample).
- Add 2 µL antibody (anti-H3K27ac antibody ABIN2668475, anti-H3K4me positive control antibody ABIN3023251, and guinea pig anti-rabbit IgG negative control antibody ABIN101961) to the respective tube, corresponding to a 1:100 dilution.
- Incubate ON at 4 °C.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Wash with 200 µL of Wash Buffer using a multichannel pipette to accelerate the process.
- Repeat the wash five times for a total of six washes.
- pAG-MNase Binding
- Prepare a 1.5 mL microcentrifuge tube containing 200 µL of pAG mix for each sample (200 µl of wash buffer + 120 ng pAG-MNase per sample).
- Place the PCR tubes with the sample on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove tubes from the magnetic stand.
- Resuspend the beads in 200 µL of pAG-MNase premix.
- Incubate for 30 min at 4 °C.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Wash with 200 µL of Wash Buffer using a multichannel pipette to accelerate the process.
- Repeat the wash for a total of five washes.
- Resuspend in 200 µL of Wash Buffer.
- MNase digestion and release of pAG-MNase-antibody-chromatin complexes
- Place PCR tubes on ice and allow to chill.
- Prepare a 1.5 mL microcentrifuge tube with 51 µl of 2 mM CaCl2 mix per sample (50 µl Wash Buffer + 1 µL 100 mM CaCl2) and let it chill on ice.
- Always in ice, place the samples on the magnetic rack and when the liquid is clear remove the supernatant.
- Resuspend the samples in 50 µl of the 2 mM CaCl2 mix and incubate in ice for exactly 30 min.
- Place the sample on the magnet stand and when the liquid is clear move the supernatant in fresh collection tubes with 3µl of EDTA/EGTA 0.25M (Digestion buffer).
- Resuspend the sample in 47 µl of 1x Urea STOP Buffer (8.5 M Urea, 100 mM NaCl, 2 mM EGTA, 2 mM EDTA, 0,5% IGEPAL).
- Incubate the samples 1 h at 4 °C.
- Transfer the supernatant containing the pAG-MNase-bound digested chromatin fragments to the previously collected digestion buffer.
- DNA Clean up
- Take the Mag-Bind® TotalPure NGS beads (Omega Bio-Tek, M1378-01) from the storage and wait until they are at RT.
- Add 2x volume of beads to each sample (e.g. 100 µL of beads for 50 µL of sample).
- Incubate the beads and the sample for 15 min at RT.
- During incubation prepare fresh EtOH 80%.
- Place the PCR tubes on a magnet stand and when the liquid is clear remove the supernatant.
- Add 200 µl of fresh 80% EtOH to the sample without disturbing the beads (Important!!! Do NOT resuspend the beads or remove the tubes from the magnet stand or the sample will be lost).
- Incubate 30 sec at RT.
- Remove the EtOH from the sample.
- Repeat the wash with 80% EtOH.
- Resuspend the beads in 25 µL of 10 mM Tris.
- Incubate the sample for 2 min at RT.
- Repeat the 2x beads clean up as described before (this time with 50 µL of beads for each sample).
- Resuspend the beads + DNA in 20 µL of 10 mM Tris.
- Library preparation and sequencing
- Prepare libraries using KAPA HyperPrep Kit using KAPA Dual-Indexed adapters according to protocol.
- Sequence samples on an Illumina NextSeq 500 sequencer, using a NextSeq 500/550 High Output Kit v2.5 (75 Cycles), 36bp PE.
- Bioinformatics
- Align reads the human genome (hg38) using bowtie78 with settings -X 700 -m1 -v 3. Remove duplicate reads, and sort files using samtools. Filter mapped reads for size, keeping only reads with a fragment size at or below 120 base pairs.
- Generate bedgraph files using bedtools genomecov.
- Call peaks using SEACR version 1.3, in relaxed mode, normalizing to the negative control.
- Experimental Notes
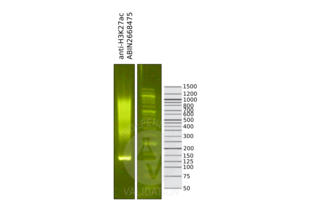
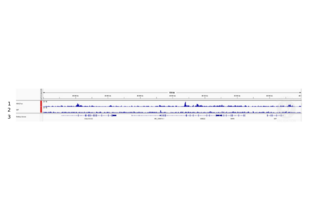
Validation #104489 (Cleavage Under Targets and Release Using Nuclease)![Successfully validated 'Independent Validation' Badge]()
![Successfully validated 'Independent Validation' Badge]() Validation ImagesFull Methods
Validation ImagesFull Methods -
- Concentration
- 1 μg/μL
- Preservative
- Sodium azide
- Precaution of Use
- This product contains Sodium azide: a POISONOUS AND HAZARDOUS SUBSTANCE which should be handled by trained staff only.
- Handling Advice
-
Avoid repeated freeze/thaw cycles and keep on ice when not in storage.
- Storage
- -20 °C
- Storage Comment
- Antibodies in solution can be stored at -20 °C for 2 years.
- Expiry Date
- 6 months
-
-
: "BMP4 preserves the developmental potential of mESCs through Ube2s- and Chmp4b-mediated chromosomal stability safeguarding." in: Protein & cell, (2022) (PubMed).
: "H3 ubiquitination by NEDD4 regulates H3 acetylation and tumorigenesis." in: Nature communications, Vol. 8, pp. 14799, (2017) (PubMed).
: "Site- and allele-specific polycomb dysregulation in T-cell leukaemia. ..." in: Nature communications, Vol. 6, pp. 6094, (2015) (PubMed).
: "Comparison of the cytotoxicity of cladribine and clofarabine when combined with fludarabine and busulfan in AML cells: Enhancement of cytotoxicity with epigenetic modulators." in: Experimental hematology, Vol. 43, Issue 6, pp. 448-61.e2, (2015) (PubMed).
: "Retinoic acid is essential for Th1 cell lineage stability and prevents transition to a Th17 cell program." in: Immunity, Vol. 42, Issue 3, pp. 499-511, (2015) (PubMed).
: "The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains." in: Genes & development, Vol. 29, Issue 14, pp. 1507-23, (2015) (PubMed).
: "Mesodermal iPSC-derived progenitor cells functionally regenerate cardiac and skeletal muscle." in: The Journal of clinical investigation, Vol. 125, Issue 12, pp. 4463-82, (2015) (PubMed).
: "Comprehensive functional annotation of 77 prostate cancer risk loci." in: PLoS genetics, Vol. 10, Issue 1, pp. e1004102, (2014) (PubMed).
: "Synergistic Anti-Tumor Activity of EZH2 Inhibitors and Glucocorticoid Receptor Agonists in Models of Germinal Center Non-Hodgkin Lymphomas." in: PLoS ONE, Vol. 9, Issue 12, pp. e111840, (2014) (PubMed).
: "An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. ..." in: Nature genetics, Vol. 46, Issue 4, pp. 364-70, (2014) (PubMed).
: "STAT3 signaling controls satellite cell expansion and skeletal muscle repair." in: Nature medicine, Vol. 20, Issue 10, pp. 1182-6, (2014) (PubMed).
: "Identification of nuclear hormone receptor pathways causing insulin resistance by transcriptional and epigenomic analysis." in: Nature cell biology, Vol. 17, Issue 1, pp. 44-56, (2014) (PubMed).
: "The Menin-Bach2 axis is critical for regulating CD4 T-cell senescence and cytokine homeostasis." in: Nature communications, Vol. 5, pp. 3555, (2014) (PubMed).
: "Systematic mapping of occluded genes by cell fusion reveals prevalence and stability of cis-mediated silencing in somatic cells. ..." in: Genome research, Vol. 24, Issue 2, pp. 267-80, (2014) (PubMed).
: "Locus-specific editing of histone modifications at endogenous enhancers." in: Nature biotechnology, Vol. 31, Issue 12, pp. 1133-6, (2013) (PubMed).
: "Global Decrease of Histone H3K27 Acetylation in ZEB1-Induced Epithelial to Mesenchymal Transition in Lung Cancer Cells." in: Cancers, Vol. 5, Issue 2, pp. 334-56, (2013) (PubMed).
: "A novel small compound SH-2251 suppresses Th2 cell-dependent airway inflammation through selective modulation of chromatin status at the Il5 gene locus." in: PLoS ONE, Vol. 8, Issue 4, pp. e61785, (2013) (PubMed).
: "A map of the cis-regulatory sequences in the mouse genome." in: Nature, Vol. 488, Issue 7409, pp. 116-20, (2012) (PubMed).
: "Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer." in: Genome research, Vol. 22, Issue 2, pp. 246-58, (2012) (PubMed).
: "Role of histone acetylation in the stimulatory effect of valproic acid on vascular endothelial tissue-type plasminogen activator expression." in: PLoS ONE, Vol. 7, Issue 2, pp. e31573, (2012) (PubMed).
-
: "BMP4 preserves the developmental potential of mESCs through Ube2s- and Chmp4b-mediated chromosomal stability safeguarding." in: Protein & cell, (2022) (PubMed).
-
- Target
- Histone 3 (H3)
- Alternative Name
- Histone H3 (H3 Products)
- Synonyms
- H-3 antibody, histocompatibility 3 antibody, H3 antibody
- Background
- Histone H3 is one of the core components of the nucleosome. The nucleosome is the smallest subunit of chromatin and consists of 147 base pairs of DNA wrapped around an octamer of core histone proteins (two each of Histone H2A, Histone H2B, Histone H3 and Histone H4). Chromatin is subject to a variety of chemical modifications, including post- translational modifications of the histone proteins and the methylation of cytosine residues in the DNA. Reported histone modifications include acetylation, methylation, phosphorylation, ubiquitylation, glycosylation, ADP-ribosylation, carbonylation and SUMOylation, these modifications play a major role in regulating gene expression. Lysine 27 of histone H3 can also be mono-, di- or trimethylated by different histone methyltransferases, such as EZH2 or NSD3. While histone methylation can be associated with transcriptional activation or repression, methylation of Lysine 27 of histone H3 is mainly associated with transcriptional repression.
- Molecular Weight
- 17 kDa
- Gene ID
- 3020
-

 (23 references)
(23 references) (1 validation)
(1 validation)



