Exosomes are extracellular microvesicles (EMV) excreted by most cells involved in intercellular communication. They are present in bodily fluids and can contain a vast array of different proteins depending on their host cell. Their content is further modulated by the cellular state such as stress or activation, or inhibition of specific signaling pathways. This makes exosomes excellent biomarkers for liquid biopsies e.g. in cancer diagnosis.
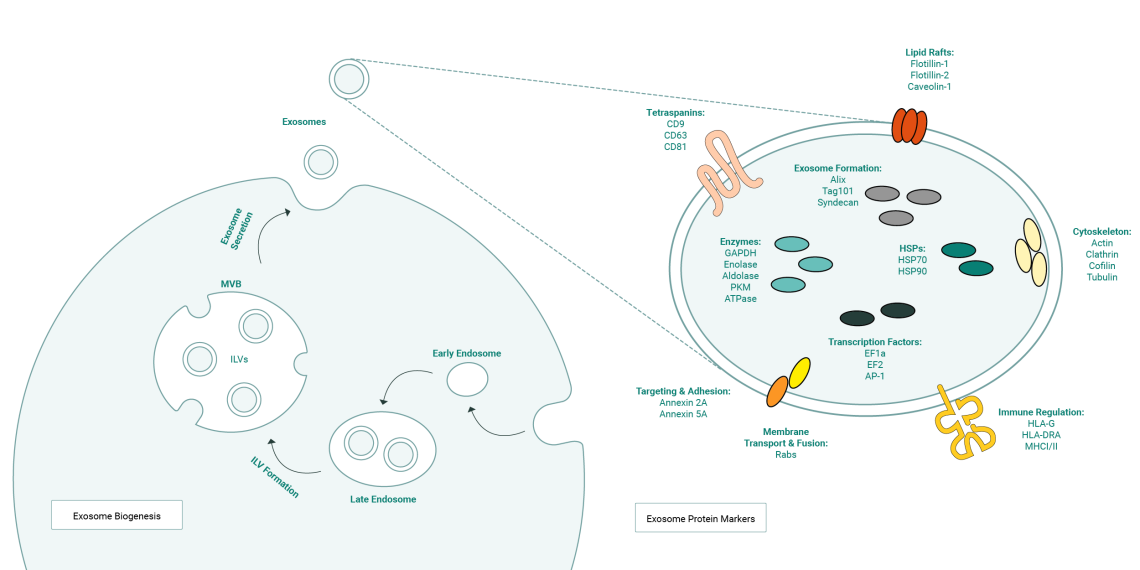
Exosome Biogenesis begins with the invagination of early endosomes. These mature into late endosomes and then further into multivesicular bodies (MVBs). MVB formation involves the inward budding of the endosomal membrane, leading to the sequestration of cytoplasmic components into intraluminal vesicles (ILVs). ILVs are released through exocytosis into the extracellular space, which earned exosomes their name. Exosomes can contain a wide variety of exosome protein markers depending on their host cell which and the cell’s state, e.g. stress or activation, or inhibition of specific signaling pathways). Tetraspanins like CD9, CD63 and CD81 are the most common canonical exosome marker proteins. Surface localization of tetraspanins makes them particularly well suited for immunolabeling and purification of exosomes from biological samples. Components of the endosomal sorting complex required for transport (ESCRT) like TSG101 and Alix, cytoskeletal proteins, integrins and annexins are also enriched on exosomes. These molecules play a pivotal role in exosome targeting and cell adhesion.
What are Exosomes?
Exosomes are small (50-120nm) endosome-derived extracellular microvesicles (EMV) that play a crucial role in intercellular communication by transporting various biomolecules, such as proteins, lipids, and nucleic acids, between cells. They were first observed in the early 1980s in the culture media of reticulocytes. The term exosome is based on the observation that they are released through exocytosis into the extracellular media. Exosomes share similar topology to the plasma membrane and are released by virtually all cell types and have been confirmed in all bodily fluids.
Get our exosome marker poster!
By clicking on the link below, you can download a copy of our Exosome poster in PDF format.
Download a CopyHow are Exosomes formed?
Exosome biogenesis usually begins with the internalization of extracellular material through endocytosis leading to the formation of early endosomes, which are membrane-bound organelles that contain the internalized cargo. These then mature into late endosomes, a process accompanied by changes in membrane composition and cargo sorting. Late endosomes can develop into MVBs, which are characterized by the presence of intraluminal vesicles (ILVs) formed through invagination of the endosomal membrane. In the MVB, proteins, lipids, nucleic acids, and other molecules are sorted into ILVs. This sorting is a critical step in exosome biogenesis and is achieved by the endosomal sorting complexes required for transport (ESCRT). The ESCRT consists of several protein complexes (ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III) that work together to recognize and sequester cargo into budding ILVs. MVBs can then either fuse with lysosomes for degradation, or they can fuse with the plasma membrane, releasing the ILVs into the extracellular space as exosomes.
How are Exosomes Identified?
Exosomes contain a vast array of different proteins depending on their host cell which, and their components are further modulated by cellular state (e.g. stress or activation, or inhibition of specific signaling pathways). Tetraspanins like CD9, CD63 and CD81 are the most common canonical exosome marker proteins, present on the vesicle surface. Surface localization of tetraspanin antigens makes them good candidate targets for immunolabeling and purification of exosomes from biological samples. Components of the endosomal sorting complex required for transport (ESCRT) like TSG101 and Alix, cytoskeletal proteins, integrins and annexins are also enriched on exosomes; these molecules play a pivotal role in exosome targeting and cell adhesion.
Why are Exosomes important?
Secretion of exosomes occurs constitutively though the rate of exosome secretion, and composition of exosomes may be augmented by a variety of intrinsic or extrinsic factors (e.g. cell stress, signaling cascades). Despite their ubiquitous nature, exosomes are considered unconventional secretory pathway components.
Because exosomes are secreted from nearly every cell type, their composition mirrors their host diversity, and depends heavily upon the type of cell from which they originate. Their molecular composition also reflects physiological or pathophysiological changes in their cell or tissue of origin. This makes exosomes excellent biomarkers for liquid biopsies to diagnose and track disease progression. For cancer diagnostics, exosomes have significant advantages over circulating tumor cells (CTCs) or circulating tumor DNA (ctDNA) because of their abundance, stability, and the wide variety of contained marker molecules. Exosomes are also implicated in cell-cell communication. Exosome components may be transferred directly to neighboring cells or may be shuttled across different cells before reaching their end destination via a method known as transcytosis. This way, exosomes can transmit signals across large distances where simple diffusion may be insufficient. Their role in cell-cell communication suggests that exosomes may have a deeper role in many physiological processes; this hypothesis is supported by the observation that exosome signaling plays a direct role in development and patterning, immune response, neuronal communication, and tissue repair. In some pathologies, exosomes also act as vectors; tumor cell-derived exosomes play an active role in tumor angiogenesis and metastasis. Exosomes shed from stimulated blood cells and the vascular endothelium are involved in neurological disorders such as multiple sclerosis, transient ischemic attacks, and antiphospholipid syndrome. Exosomes may also carry damaged cellular material targeted for destruction and facilitate the spreading of toxic forms of aggregated proteins such as α-synuclein and β-amyloid and contribute to the progression of neurodegenerative diseases. Some research also suggests that exosome transport has been exploited by viral pathogens such as SARS-CoV-2 to travel between host cells and evade immune detection. Because of their small size and simple structure, exosomes may sometimes cross the blood-brain barrier. It has been suggested that exosomes could serve as a delivery system targeting the central nervous system to treat neuropathic diseases without the need for invasive surgery. The use of exosomes to transfer genetic information, or to deliver therapeutic agents is a currently underexplored field that holds vast medicinal potential.How are Exosomes Studied?
The exosome secretome is vast and diverse, containing many different markers (see http://www.exocarta.org/). The presence of canonical surface markers like those listed above permits purification and in-depth study of exosome secretion and content from different sample types.
antibodies-online offers a range of antibodies and ELISA kits for the detection of know exosome proteins.
Exosome Marker Antibodies
Based on recent literature, the most relevant protein exosome markers include:
| Protein | Gene | GeneID | Uniprot | Ref | exocarta Top 100 proteins | TS | EF | LP | TA | CS | AG | MT | AP | HS | EN | RG | CA | II | VI | ND |
| 14-3-3 protein epsilon | YWHAE | 7531 | P62258 | - | 22 | X | ||||||||||||||
| 14-3-3 protein beta/alpha | YWHAB | 7529 | P31946 | 50 | x | |||||||||||||||
| 14-3-3 protein eta | YWHAH | 7533 | Q04917 | 94 | x | |||||||||||||||
| 14-3-3 protein gamma | YWHAG | 7532 | P61981 | 54 | x | |||||||||||||||
| 14-3-3 protein theta | YWHAQ | 10971 | P27348 | 56 | x | |||||||||||||||
| 14-3-3 protein zeta/delta | YWHAZ | 7534 | P63104 | - | 15 | X | ||||||||||||||
| 78 kDa glucose-regulated protein | HSPA5 | 3309 | P11021 | (1) | 35 | X | ||||||||||||||
| Actin, cytoplasmic 1 | ACTB | 60 | P60709 | - | 5 | X | ||||||||||||||
| ADAM10 | ADAM10 | 102 | O14672 | (2) | - | X | X | X | X | |||||||||||
| Alix | PDCD6IP | 10015 | Q8WUM4 | (3) | 2 | X | ||||||||||||||
| Alpha-Enolase | ENO1 | 2023 | P06733 | (4) | 9 | X | ||||||||||||||
| Alpha-Synclein | SNCA | 6622 | P37840 | (32) | - | X | X | |||||||||||||
| Aminopeptidase N | ANPEP | 290 | P15144 | - | - | X | X | |||||||||||||
| Beta-amyloid | APP | 351 | P05067 | (5) | - | X | ||||||||||||||
| Annexin A1 | ANXA1 | 301 | P04083 | 53 | x | |||||||||||||||
| Annexin A11 | ANXA11 | 311 | P50995 | 68 | x | |||||||||||||||
| Annexin A4 | ANXA4 | 307 | P09525 | 72 | x | |||||||||||||||
| Annexin A6 | ANXA6 | 309 | P08133 | 67 | x | |||||||||||||||
| Annexin A5 | ANXA5 | 308 | P08758 | (6) | 20 | X | X | |||||||||||||
| Annexin A2 | ANXA2 | 302 | P07355 | (7) | 6 | X | ||||||||||||||
| AP-1 | JUN | 3725 | P05412 | - | - | X | X | |||||||||||||
| ATP citrate lyase | ACLY | 47 | P53396 | - | 72 | X | ||||||||||||||
| ATPase | ATP1A1 | 476 | P05023 | - | 39 | X | ||||||||||||||
| Basigin | BSG | 682 | P35613 | (8) | - | X | X | |||||||||||||
| Caveolin-1 | CAV1 | 857 | Q03135 | (9), (10) | - | X | X | |||||||||||||
| CD9 | CD9 | 928 | P21926 | (11) | 1 | X | ||||||||||||||
| CD11a | ITGAL | 3683 | P20701 | (12) | - | X | X | |||||||||||||
| CD11b | ITGAX | 3687 | P11215 | (12) | - | X | X | |||||||||||||
| CD11c | ITGAM | 3684 | P20702 | (12) | - | X | X | |||||||||||||
| CD29 | ITGB1 | 3688 | P05556 | (12) | 34 | X | X | |||||||||||||
| CD37 | CD37 | 951 | P11049 | (11) | - | X | ||||||||||||||
| CD44 | CD44 | 960 | P16070 | (13) | - | X | X | |||||||||||||
| CD49f | ITGA6 | 3655 | P23229 | (12) | 89 | X | X | |||||||||||||
| CD53 | CD53 | 963 | P19397 | - | x | |||||||||||||||
| CD63 | CD63 | 967 | P08962 | (10), (11) | 7 | X | X | |||||||||||||
| CD81 | CD81 | 975 | P60033 | (14) | 24 | X | X | |||||||||||||
| CD82 | CD82 | 3732 | P27701 | (11) | - | X | ||||||||||||||
| CD142 | TF | 2152 | P13726 | (15) | - | X | X | |||||||||||||
| CD146 | MCAM | 4162 | P43121 | (15) | - | X | X | |||||||||||||
| CD163 | CD163 | 9332 | Q86VB7 | (15) | - | X | X | X | ||||||||||||
| Clathrin heavy chain 1 | CLTC | 1213 | Q00610 | - | 23 | X | ||||||||||||||
| Claudin-1 | CLDN1 | 9076 | O95832 | (8) | - | X | ||||||||||||||
| Cofilin-1 | CFL1 | 1072 | P23528 | - | 25 | X | ||||||||||||||
| - | - | (16) | - | X | ||||||||||||||||
| - | - | (16) | - | X | ||||||||||||||||
| EF-1-alpha-1 | EEF1A1 | 1915 | P68104 | (4) | 14 | X | ||||||||||||||
| EF2 | EEF2 | 1938 | P13639 | - | 17 | X | ||||||||||||||
| EGFR | EGFR | 1956 | P00533 | (15) | - | X | ||||||||||||||
| Ep-CAM | EPCAM | 4072 | P16422 | (17), (18) | - | X | X | |||||||||||||
| Fatty acid synthase | FASN | 2194 | P49327 | (3) | 21 | X | X | |||||||||||||
| Fibronectin | FN1 | 2335 | P02751 | 93 | ||||||||||||||||
| Flotillin-1 | FLOT1 | 10211 | O75955 | (18), (19) | 41 | X | X | |||||||||||||
| Flotillin-2 | FLOT2 | 2319 | Q14254 | (19) | - | X | ||||||||||||||
| Fructose-bisphosphate aldolase A | ALDOA | 226 | P04075 | - | 18 | X | ||||||||||||||
| Gelsolin | GSN | 2934 | P06396 | 92 | x | x | ||||||||||||||
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 2597 | P04406 | - | 4 | X | ||||||||||||||
| HCV core protein | - | - | (20) | - | X | |||||||||||||||
| Heat shock 70 kDa protein 1A | HSPA1A | 3303 | P0DMV8 | 51 | x | |||||||||||||||
| Heat shock protein HSP 90-alpha | HSP90AA1 | 3320 | P07900 | (1) | 10 | X | X | |||||||||||||
| Heat shock protein HSP 90-beta | HSP90AB1 | 3326 | P08238 | (1) | 19 | X | ||||||||||||||
| Heparanase | HPSE | 10855 | Q9Y251 | (21) | - | X | X | |||||||||||||
| - | - | (20) | - | X | ||||||||||||||||
| - | - | (20) | - | X | ||||||||||||||||
| HLA-DRA | HLA-DRA | 3122 | P01903 | (22) | - | X | X | X | ||||||||||||
| HLA-G | 3135 | P17693 | (23) | - | X | X | X | |||||||||||||
| Hsc70 | HSPA8 | 3312 | P11142 | (1) | 3 | X | ||||||||||||||
| - | - | (16) | - | X | ||||||||||||||||
| Tax | - | - | (20) | - | X | |||||||||||||||
| Huntingtin | HTT | 3064 | P42858 | (5) | - | X | X | |||||||||||||
| ICAM-1 | ICAM1 | 3383 | P05362 | (24) | - | X | ||||||||||||||
| Leucine-rich receptor kinase 2 | LRRK2 | 120892 | Q5S007 | (5) | - | X | ||||||||||||||
| L-lactate dehydrogenase A chain | LDHA | 3939 | P00338 | - | 13 | X | ||||||||||||||
| Lysosome-associated membrane glycoprotein 1 | LAMP1 | 3916 | P11279 | (25) | - | X | X | |||||||||||||
| Lysosome-associated membrane glycoprotein 2 | LAMP2 | 3920 | P13473 | (23) | 88 | X | X | |||||||||||||
| MHCI | - | - | (26) | - | X | X | ||||||||||||||
| MHCII | - | - | (26) | - | X | X | ||||||||||||||
| MUC1 | MUC1 | 4582 | P15941 | (15) | - | X | X | |||||||||||||
| N-cadherin | CDH2 | 1000 | P19022 | (15) | - | X | X | |||||||||||||
| Phosphoglycerate kinase 1 | PGK1 | 5230 | P00558 | (4) | 16 | X | ||||||||||||||
| Placental Alkaline Phosphatase | ALPP | 250 | P05187 | (15) | - | X | X | |||||||||||||
| Prion proteins | - | - | (5) | - | X | |||||||||||||||
| Prostate-specific antigen | KLK3 | 354 | P07288 | (27) | - | X | X | |||||||||||||
| Pyruvate kinase PKM | PKM | 5315 | P14618 | (28) | 12 | X | X | |||||||||||||
| Rab-14 | RAB14 | 51552 | P61106 | - | 75 | X | ||||||||||||||
| Rab-5a | RAB5A | 5868 | P20339 | - | 80 | X | ||||||||||||||
| Rab-5b | RAB5B | 5869 | P61020 | - | 86 | X | ||||||||||||||
| Rab-5c | RAB5C | 5878 | P51148 | - | 64 | X | ||||||||||||||
| Rab-7 | RAB7A | 7879 | P51149 | - | 61 | X | ||||||||||||||
| Rap 1B | RAP1B | 5908 | P61224 | - | 33 | X | ||||||||||||||
| Superoxide dismutase | SOD1 | 6647 | P00441 | - | x | x | ||||||||||||||
| Syndecan-1 | SDC1 | 6382 | P18827 | (29) | - | X | ||||||||||||||
| Syndecan-4 | SDC4 | 6385 | P31431 | (29) | - | X | ||||||||||||||
| Syntenin-1 | SDCBP | 6386 | O00560 | (30) | 8 | X | ||||||||||||||
| TARDBP | TDP-43 | 23435 | Q13148 | - | x | |||||||||||||||
| Transitional endoplasmic reticulum ATPase | VCP | 7415 | P55072 | 26 | ||||||||||||||||
| Triosephosphate isomerase | TPI1 | 7167 | P60174 | 27 | x | |||||||||||||||
| Tumor-Associated Glycoprotein | TAG-72 | - | - | (15) | - | X | ||||||||||||||
| Tetraspanin-8 | Tspan8 | 7103 | P19075 | (15) | - | X | X | |||||||||||||
| Tsg101 | TSG101 | 7251 | Q99816 | (31) | 11 | X | ||||||||||||||
| Tubulin alpha-1C chain | TUBA1C | 84790 | Q9BQE3 | - | x | |||||||||||||||
| Tubulin alpha-4A chain | TUBA4A | 7277 | P68366 | - | x | |||||||||||||||
| Tubulin beta-2B chain | TUBB2B | 347733 | Q9BVA1 | - | x | |||||||||||||||
| Tubulin beta-4B chain | TUBB4B | 10383 | P68371 | - | x | |||||||||||||||
| Vacuolar-sorting protein 35 | VPS35 | 55737 | Q96QK1 | (5) | - | X | X |

Get our exosome marker poster!
By clicking on the link below, you can download a copy of our Exosome poster in PDF format.
Download a CopyReferences
- : "ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming." in: Blood, Vol. 106, Issue 1, pp. 216-23, (2005) (PubMed).
- : "Exosome-dependent trafficking of HSP70: a novel secretory pathway for cellular stress proteins." in: The Journal of biological chemistry, Vol. 280, Issue 24, pp. 23349-55, (2005) (PubMed).
- : "The regulation of exosome secretion: a novel function of the p53 protein." in: Cancer research, Vol. 66, Issue 9, pp. 4795-801, (2006) (PubMed).
- : "B cell activation regulates exosomal HLA production." in: European journal of immunology, Vol. 38, Issue 5, pp. 1423-34, (2008) (PubMed).
- : "CD44 and EpCAM: cancer-initiating cell markers." in: Current molecular medicine, Vol. 8, Issue 8, pp. 784-804, (2009) (PubMed).
- : "High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients." in: PloS one, Vol. 4, Issue 4, pp. e5219, (2009) (PubMed).
- : "Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer." in: BMC cancer, Vol. 9, pp. 244, (2009) (PubMed).
- : "Lipid raft endocytosis and exosomal transport facilitate extracellular trafficking of annexin A2." in: The Journal of biological chemistry, Vol. 286, Issue 35, pp. 30911-30925, (2011) (PubMed).
- : "Microvesicles and viral infection." in: Journal of virology, Vol. 85, Issue 24, pp. 12844-54, (2012) (PubMed).
- : "The roles of flotillin microdomains--endocytosis and beyond." in: Journal of cell science, Vol. 124, Issue Pt 23, pp. 3933-40, (2012) (PubMed).
- : "Soluble serum CD81 is elevated in patients with chronic hepatitis C and correlates with alanine aminotransferase serum activity." in: PloS one, Vol. 7, Issue 2, pp. e30796, (2012) (PubMed).
- : "Syndecan-syntenin-ALIX regulates the biogenesis of exosomes." in: Nature cell biology, Vol. 14, Issue 7, pp. 677-85, (2012) (PubMed).
- : "Quantitative proteomic analysis of exosomes from HIV-1-infected lymphocytic cells." in: Proteomics, Vol. 12, Issue 13, pp. 2203-11, (2012) (PubMed).
- : "Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy." in: Human molecular genetics, Vol. 21, Issue R1, pp. R125-34, (2013) (PubMed).
- : "Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus." in: American journal of respiratory cell and molecular biology, Vol. 48, Issue 6, pp. 742-8, (2013) (PubMed).
- : "Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes." in: The Journal of biological chemistry, Vol. 288, Issue 14, pp. 10093-10099, (2013) (PubMed).
- : "Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1." in: The Journal of biological chemistry, Vol. 288, Issue 24, pp. 17713-24, (2013) (PubMed).
- : "Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer." in: PloS one, Vol. 8, Issue 12, pp. e82589, (2014) (PubMed).
- : "Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2." in: Nature communications, Vol. 5, pp. 3477, (2014) (PubMed).
- : "Plasma exosomal α-synuclein is likely CNS-derived and increased in Parkinson's disease." in: Acta neuropathologica, Vol. 128, Issue 5, pp. 639-650, (2015) (PubMed).
- : "Tetraspanins in extracellular vesicle formation and function." in: Frontiers in immunology, Vol. 5, pp. 442, (2014) (PubMed).
- : "Exosomes secreted by cortical neurons upon glutamatergic synapse activation specifically interact with neurons." in: Journal of extracellular vesicles, Vol. 3, pp. 24722, (2014) (PubMed).
- : "Antigen Presentation by MHC-Dressed Cells." in: Frontiers in immunology, Vol. 5, pp. 672, (2015) (PubMed).
- : "Anti-cancer fatty-acid derivative induces autophagic cell death through modulation of PKM isoform expression profile mediated by bcr-abl in chronic myeloid leukemia." in: Cancer letters, Vol. 360, Issue 1, pp. 28-38, (2015) (PubMed).
- : "Exosomes: novel biomarkers for clinical diagnosis." in: TheScientificWorldJournal, Vol. 2015, pp. 657086, (2016) (PubMed).
- : "Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma." in: Journal of extracellular vesicles, Vol. 4, pp. 26659, (2015) (PubMed).
- : "HLA-G: An Immune Checkpoint Molecule." in: Advances in immunology, Vol. 127, pp. 33-144, (2015) (PubMed).
- : "Exosomes and Their Role in the Life Cycle and Pathogenesis of RNA Viruses." in: Viruses, Vol. 7, Issue 6, pp. 3204-25, (2016) (PubMed).
- : "ExoCarta: A Web-Based Compendium of Exosomal Cargo." in: Journal of molecular biology, Vol. 428, Issue 4, pp. 688-692, (2016) (PubMed).
- : "Tumour exosome integrins determine organotropic metastasis. ..." in: Nature, Vol. 527, Issue 7578, pp. 329-35, (2015) (PubMed).
- : "Exosomes in neurological disease, neuroprotection, repair and therapeutics: problems and perspectives." in: Neural regeneration research, Vol. 10, Issue 10, pp. 1565-7, (2015) (PubMed).
- : "Interrogating Circulating Microsomes and Exosomes Using Metal Nanoparticles." in: Small (Weinheim an der Bergstrasse, Germany), Vol. 12, Issue 6, pp. 727-32, (2016) (PubMed).
- : "PSA and beyond: alternative prostate cancer biomarkers." in: Cellular oncology (Dordrecht), Vol. 39, Issue 2, pp. 97-106, (2016) (PubMed).
- : "A Perspective on Extracellular Vesicles Proteomics." in: Frontiers in chemistry, Vol. 5, pp. 102, (2017) (PubMed).
- : "Reassessment of Exosome Composition." in: Cell, Vol. 177, Issue 2, pp. 428-445.e18, (2019) (PubMed).
- : "On the potential role of exosomes in the COVID-19 reinfection/reactivation opportunity." in: Journal of biomolecular structure & dynamics, Vol. 39, Issue 15, pp. 5831-5842, (2021) (PubMed).
- : "Ultrastructural Evidence for Direct Renal Infection with SARS-CoV-2." in: Journal of the American Society of Nephrology : JASN, Vol. 31, Issue 8, pp. 1683-1687, (2020) (PubMed).
- : "Extracellular vesicles and amyotrophic lateral sclerosis: from misfolded protein vehicles to promising clinical biomarkers." in: Cellular and molecular life sciences : CMLS, Vol. 78, Issue 2, pp. 561-572, (2021) (PubMed).
- : "The exosome journey: from biogenesis to uptake and intracellular signalling." in: Cell communication and signaling : CCS, Vol. 19, Issue 1, pp. 47, (2022) (PubMed).
- : "Emerging Role of Exosomes in Liquid Biopsy for Monitoring Prostate Cancer Invasion and Metastasis." in: Frontiers in cell and developmental biology, Vol. 9, pp. 679527, (2021) (PubMed).
- : "Valosin-Containing Protein (VCP)/p97: A Prognostic Biomarker and Therapeutic Target in Cancer." in: International journal of molecular sciences, Vol. 22, Issue 18, (2021) (PubMed).
- : "Exosomes as a new frontier of cancer liquid biopsy." in: Molecular cancer, Vol. 21, Issue 1, pp. 56, (2022) (PubMed).
- : "Gelsolin: A comprehensive pan-cancer analysis of potential prognosis, diagnostic, and immune biomarkers." in: Frontiers in genetics, Vol. 14, pp. 1093163, (2023) (PubMed).
- : "Vesiclepedia 2024: an extracellular vesicles and extracellular particles repository." in: Nucleic acids research, Vol. 52, Issue D1, pp. D1694-D1698, (2024) (PubMed).
- : "Roles of exosomes in immunotherapy for solid cancers." in: Cell death & disease, Vol. 15, Issue 2, pp. 106, (2024) (PubMed).





