Histone Code
Categories
Histone proteins form the protein core of the nucleosomes, the basic organizational unit of chromatin in eukaryotes. Addition of certain chemical groups to amino acids outside of the histone protein core impacts the packaging of chromatin. This specific pattern of chemical modifications on histone proteins influences gene expression by controlling the accessibility of DNA to the transcriptional machinery. It is referred to as the histone code. Together with similar modifications such as DNA methylation it is part of the epigenetic code.
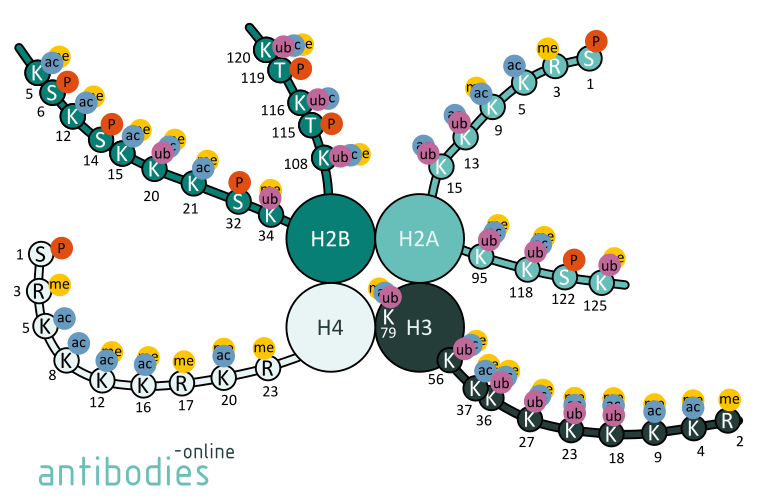
Fig. 1: Examples for histone modifications of selected amino acid positions in histones H2A, H2B, H3, and H4.
Post-translational modifications (PTMs) like methylation or acetylation serve as epigenetic marking system that recruits other proteins by specific recognition of the modified histone via protein domains specialized for such purposes, rather than through simply stabilizing or destabilizing the interaction between histone and the underlying DNA. These recruited proteins then act to alter chromatin structure actively and to promote or suppress transcription.
Histone Modifications and their Impact
Histone modifications are covalent PTM of histone proteins. They act in diverse biological processes such as transcriptional activation/inactivation, chromosome packaging, and DNA damage/repair.
For instance, mono- and tri-methylated histone H3K4 are conducive to gene transcription. H3K4me3 is typically enriched at active promoters near transcription start sites. Likewise, H3K9ac is a mark for active promoters. Tri-methylation of Lysine 27 of histone 3 is characteristic for polycomb silencing through the polycomb repressive complex whereas acetylation of H3K27 is considered to be a transcription enhancer. H3K36me3 prevents aberrant initiation of transcription. It is also implicated in DNA damage response and recruiting components of the DNA damage repair machinery. Deregulation of these epigenetic patterns can severely effect protein expression and the alterations in epigenetic patterns are indeed observed in many cancers.
Epigenetic changes offer the unique possibility to alter gene expression in a reversible way and are considered a gateway to new therapy approaches. Understanding alterations has the potential to explain mechanisms of aging, human development, and the origins of several health conditions, e.g. cancer, heart disease and mental illness.
Histone Acetylation
In general histone acetylation determine the histone assembling as well as the folding and compactness of the DNA-histone interaction and therefore presenting a switch between condensed and uncondensed chromatin structure. The latter, known as euchromatin, is transcriptionally active, whereas the former, known as heterochromatin, is transcriptionally inactive.
Histone acetyltransferases (HAT) transfer an acetyl group to specific lysine residues on histones, usually residues in the N-terminal tail. Acetylated lysine side chains of histones lose their positive charge and thus the ability to form salt bridges with the negatively charged phosphate backbone of DNA. H3 and H4 histone proteins are the primary targets of HATs.
Acetylation of H3K56, H3K64 and H3K122 or H4K16 disrupts the chromatin structure and results in lightly packed euchromatin. Euchromatin participates in the active transcription of DNA. The unfolded structure allows gene regulatory proteins and RNA polymerase complexes to bind to the DNA sequence and consequently leads to an increase of gene transcription.
Histone H3K27me3 - shutting down Transcription
H3K27me3 is a deeply studied target of epigenetic researchers looking for inactive genes. Unlike other histone methylations, H3K27m3 has only one known methyltransferase: EZH2. EZH2 is responsible for the repression many genes involved in development and cell differentiation. Therefore H3K27m3 is critical for the repression of developmental genes acting in opposition to H3K4m3. When H3K27 is trimethylated, it is tightly associated with inactive gene promoters.
DNA and histone lysine methylation systems are highly interrelated and rely mechanistically on each other for normal chromatin function in vivo. Non-methylated DNA in CpG islands can act as part of a genomic signature to recruit H3K4 and H3K27 trimethylation, and to exclude H3K36 methylation, possibly creating chromatin environments unique to gene regulatory elements that are able to modulate transcriptional states. Functional links between 5-hydroxymethylcytosine and histone lysine methylation are in focus of current research, for example the ability of Uhrf1 and MeCP2 to als bind to 5hmC.
The mono- and di-methylation states are less studied; however, H3K27m2 shows a similar distribution to H3K27, while H3K27m1 is associated with active promoters. H3K27 can also be target of acetylation. Lysine residues can only be methylated or acetylated, in comparison the acetylation leads to opposite effects - with active transcription and antagonism of H3K27me3 regulated genes.
Histone Methylation as Therapeutic Target
Dysregulation of histone lysine methylation has been implicated in several cancers and developmental defects. Therefore, histone lysine methylation has been considered a potential therapeutic target, and clinical trials of several inhibitors of this process have shown promising results. A more detailed understanding of histone lysine methylation is necessary for elucidating complex biological processes and, ultimately, for developing and improving disease treatments. Several approaches are viable to study histones, including chromatin immunoprecipitation (together with its large-scale variants ChIP-on-chip and ChIP-Seq), fluorescent in situ hybridization or bisulfite sequencing. Recently cleavage under targets and release using nuclease (CUT&RUN) gained more and more traction as a consequent improvement for ChIP based methods.
antibodies-online offers a wide range of antibodies and for your epigenetic research needs. Browse our portfolio down below! If you have any questions, our team of biologists is at your service at all times via chat, contact form or e-mail.
References
: "The language of covalent histone modifications." in: Nature, Vol. 403, Issue 6765, pp. 41-5, (2000) (PubMed).: "Understanding the relationship between DNA methylation and histone lysine methylation." in: Biochimica et biophysica acta, Vol. 1839, Issue 12, pp. 1362-72, (2015) (PubMed).
: "Histone methylation and the DNA damage response." in: Mutation research. Reviews in mutation research, Vol. 780, pp. 37-47, (2020) (PubMed).
: "Independent manipulation of histone H3 modifications in individual nucleosomes reveals the contributions of sister histones to transcription." in: eLife, Vol. 6, (2018) (PubMed).

Goal-oriented, time line driven scientist, proficiently trained in different academic institutions in Germany, France and the USA. Experienced in the life sciences e-commerce environment with a focus on product development and customer relation management.
Go to author page



