Magnetic Concanavalin A Beads (Agarose)
-
- Key Features
-
- Magnetic agarose concanavalin A beads for CUT&RUN and CUT&Tag assays.
- Superior handling compared to magnetic silica concanavalin A beads: easier washes and resuspension.
- Reduced risk for samples to dry out.
- Performance comparable to or better than magnetic silica concanavalin A beads.
- Application
- Cleavage Under Targets and Release Using Nuclease (CUT&RUN), Cleavage Under Targets and Tagmentation (CUT&Tag), Separation (Sep)
- Purpose
- Immobilization of whole cells through Concanavlin A binding of membrane glyocproteins.
- Characteristics
-
- Magnetic ConA Beads (Agarose) for CUT&RUN/CUT&Tag Assays are based on a ferrimagnetic core surrounded by an agarose matrix covalently bound via polyurethane links.
- In contrast to silica based beads containing superparamagnetic magnetite nanaoparticles, the hydrophilic surface of our Magnetic ConA Beads (Agarose) for CUT&RUN/CUT&Tag Assays reduces the risk of unspecific binding of contaminants. No residual charges are present after conjugation. This minimizes non-specific binding to the matrix.
- The weak magnetic moment does not interfere with their solubility in the absence of an external magnet field. Upon exposure to a magnetic field however, the beads show a stronger magnetic reaction than the superparamagnetic beads. They are therefore easier to pull out of a solution using a magnetic separator.
- The beads' diameter is with 30 μm considerably larger than for silica based ConA beads. Their capacity is comparable because of the open structure of the agarose carbohydrate network.
- Components
- 10% suspension of Concanavalin A coated agarose particles with a ferrimagnetic core
- Bead Ligand
- Concanavalin A
- Bead Matrix
- Magnetic Agarose beads
- Bead Size
- 30 μm
-
-
- Application Notes
- Optimal working dilution should be determined by the investigator.
- Comment
-
- Metal ions (calcium and manganese) mediate the binding to Concanavalin A and stabilize is conformation.
- The use of buffers with EDTA or other metal chelators must be avoided as it will result in a loss of carbohydrate binding ability.
- Protocol
-
- Collect approximately 250,000 cells for each sample.
- Wash cells 3 times in 1 mL Wash Buffer (20 mM HEPES pH 7.5, 150 mM NaCl).
- Take cells up in a volume of Wash Buffer corresponding to 250,000 cells/mL.
- Homogenize Magnetic ConA Beads (Agarose) for CUT&RUN/CUT&Tag Assays slurry by shaking.
- Take 10 μL bead slurry for each sample.
- Wash beads 3 times with 1 mL Binding Buffer (20 mM HEPES pH 7.5, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2.
- Resuspend beads in a volume of Binding Buffer corresponding to the initial volume of bead slurry.
- Add beads in Binding Buffer to the cells in Wash Buffer.
- Incubate for 30 min at RT to bind cells to Magnetic ConA Beads (Agarose) for CUT&RUN/CUT&Tag Assays.
- Restrictions
- For Research Use only
-
- by
- Max Planck Institut für Immunbiologie und Epigenetik
- No.
- #104288
- Date
- 08/24/2020
- Antigen
- ConA
- Lot Number
- cab0304001
- Method validated
- Cleavage Under Targets and Release Using Nuclease
- Positive Control
- Anti-H3K4me3 antibody
- Negative Control
- guinea pig anti-rabbit antibody ABIN101961
- Notes
Passed. ABIN6952467 allows immobilization of cells for CUT&RUN.
- Primary Antibody
- Secondary Antibody
- Full Protocol
- Cell harvest
- Harvest 5,000 murine LSK cells per sample to be used at RT.
- Centrifuge cell solution 3 min at 600 x g at RT.
- Remove the liquid carefully.
- Gently resuspend cells in 1 mL Wash Buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, Roche Complete Protease Inhibitor EDTA-free) by pipetting and transfer cell solution to a 2 mL microcentrifuge tube.
- Centrifuge cell solution 3 min at 600 x g at RT and discard the supernatant.
- Repeat twice for a total of three washes.
- Resuspend cell pellet in 1 mL Wash Buffer by gently pipetting.
- Concanavalin A beads preparation
- Prepare one 1.5 mL microcentrifuge tube.
- Gently resuspend the magnetic silica-based magnetic ConA beads ABIN6923139 or agarose-based magnetic ConA beads ABIN6952467.
- Pipette 10 µL Con A Beads slurry for each sample into the 1.5 mL microcentrifuge tube.
- Place the tube on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tube from the magnetic stand.
- Pipette 1 mL Binding Buffer (20 mM HEPES pH 7.5, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2) into each tube and resuspend ConA beads by gentle pipetting.
- Spin down the liquid from the lid with a quick pulse in a table-top centrifuge.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tube from the magnetic stand.
- Repeat twice for a total of three washes.
- Gently resuspend the ConA Beads in a volume of Binding Buffer corresponding to the original volume of bead slurry, i.e. 10 µL per sample.
- Cell immobilization – binding to Concanavalin A beads
- Carefully vortex the cell suspension and add 10 µL of the Con A beads in Binding Buffer to the cell suspension for each sample.
- Close tube tightly and rotate for 10 min at RT.
- Cell permeabilization and antibody binding
- Divide cell suspension into separate 2 mL microcentrifuge tubes, one for each antibody (5,000 cells per sample).
- Place the microcentrifuge tubes on a magnetic stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Place each tube at a low angle on the vortex mixer set to a low speed and add 100 µL Digitonin Wash buffer (wash buffer with 0.025% (wt/vol) Digitonin) supplemented with 2 mM EDTA.
- Gently vortex the microcentrifuge tubes until the beads are resuspended.
- For the positive control, add 1 µL anti-H3K4me3 antibody to the respective tube, corresponding to a 1:100 dilution.
- For the negative control, add 1 µL guinea pig anti rabbit negative control antibody (antibodies-online, ABIN101961, lot NE-200-12190001) to the respective tube, corresponding to a 1:100 dilution.
- Rotate the microcentrifuge tubes ON at 4 °C.
- Spin down the liquid and place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Resuspend with 1 mL Digitonin Wash Buffer and mix by inversion. If clumping occurs, gently remove the clumps with a 1 ml pipette tip.
- Repeat once for a total of two washes.
- pA-MNase Binding
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Vortex the sample at low speed and add 150 μL pA-MNase solution at 700 ng/mL per sample, gently resuspending the beads by pipetting.
- Rotate the microcentrifuge tubes for 1 h at 4 °C.
- Spin down the liquid and place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Resuspend with 1 mL Digitonin Wash Buffer and mix by inversion. If clumping occurs, gently remove the clumps with a 1 mL pipette tip.
- Repeat once for a total of two washes.
- MNase digestion and release of pA-MNase-antibody-chromatin complexes
- Spin down the liquid from the lid with a quick pulse in a table-top centrifuge.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Place each tube at a low angle on the vortex mixer set to a low speed and add 100 μL Digitonin Wash buffer per sample along the side of the tube.
- Place tubes in a heat block, kept on ice, and allow to chill.
- Add 2 μL 0.1 M CaCl2 to each sample.
- Incubate tubes at 0 °C for 30 min.
- Add 100 μL 2xSTOP buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 0.05% (wt/vol) Digitonin, 100 μg/mL RNAse A, 50 μg/mL Glycogen).
- Incubate tubes at 37 °C for 30 min.
- Place the tubes on a magnet stand until the fluid is clear.
- Transfer the supernatant containing the pA-MNase-bound digested chromatin fragments to fresh 1.5 mL microcentrifuge tubes.
- DNA extraction
- Add 2 µL 10% SDS to a final concentration of 0.1% and 2.5 µL Proteinase K (20 mg/mL) to each supernatant.
- Gently vortex tubes at a low speed of approximately 1,100 rpm.
- Incubate tubes at 50 °C for 1 h.
- Add 200 µL PCI to tube.
- Vortex tubes thoroughly at high speed until the liquid appears milky.
- Centrifuge tubes in a tabletop centrifuge at 16,000 x g at RT for 5 min.
- Carefully transfer the upper aqueous phase to a fresh 1.5 mL microcentrifuge tube containing 200 µL chloroform:isoamyl alcohol 24:1.
- Vortex tubes thoroughly at high speed until the liquid appears milky.
- Centrifuge tubes in a tabletop centrifuge at 16,000 x g at 4 °C for 5 min.
- Carefully transfer to upper aqueous phase to a fresh 1.5 mL microcentrifuge tube containing 2 µL glycogen (diluted 1:10 to 2 mg/mL from the 20 mg/mL stock solution).
- Add 20 µL 3 M NaOAc pH 5.2.
- Add 400 µL 100% ethanol.
- Place tubes for at -20 °C ON.
- Centrifuge tubes in a tabletop centrifuge at 16,000 x g at 4 °C for 5min.
- Remove the liquid carefully with a pipette.
- Wash pellet with 1ml 70% ethanol.
- Centrifuge tubes in a tabletop centrifuge at 16,000 x g at 4 °C for 1 min.
- Remove the liquid carefully with a pipette.
- Air-dry the pellet, then dissolve in 30 µL 1 mM Tris-HCl, 0.1 mM EDTA.
- Library preparation and sequencing
- Read mapping and Peak calling
- Experimental Notes
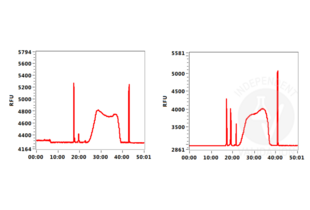
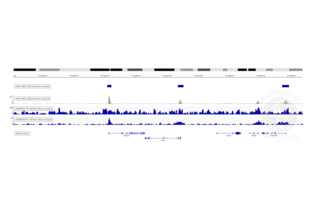
Transcription start sites were identified through CUT&RUN on LSK cells immobilized on magnetic ConA beads ABIN6952467 using an H3K4me3 antibody. Identified peaks were consistent with the Histone Mods by ChIP-seq from ENCODE (NCBI37/mm9).
Background signal using the agarose based ConA beads ABIN6952467 appeared to be lower than in a parallel experiment using silica based ConA beads for cell immobilization.
Validation #104288 (Cleavage Under Targets and Release Using Nuclease)![Successfully validated 'Independent Validation' Badge]()
![Successfully validated 'Independent Validation' Badge]() Validation ImagesFull Methods
Validation ImagesFull Methods -
- Format
- Liquid
- Buffer
- 20 mM Sodium Acetate pH 6.6, 20% Ethanol
- Handling Advice
-
Do not freeze the Magnetic ConA Beads (Agarose) for CUT&RUN/CUT&Tag Assays!
Vortex bead suspension well before use. - Storage
- 4 °C
- Expiry Date
- 12 months
-
-
: "A new cut&run low volume-urea (LoV-U) protocol optimized for transcriptional co-factors uncovers Wnt/b-catenin tissue-specific genomic targets." in: Development (Cambridge, England), (2022) (PubMed).
-
: "A new cut&run low volume-urea (LoV-U) protocol optimized for transcriptional co-factors uncovers Wnt/b-catenin tissue-specific genomic targets." in: Development (Cambridge, England), (2022) (PubMed).
-

 (1 reference)
(1 reference) (1 validation)
(1 validation)



