Hedgehog Signaling
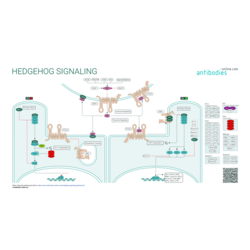
Hedgehog (Hh) signaling is highly conserved across chordates of different taxonomical classification. It is essential for development of the embryo. The Hedehog pathway was first described and characterized in the fruit fly, Drosophila melanogaster. In mammals, Hh signaling regulates cell-fate, tissue polarity, and patterning during early embryogenesis and the morphogenesis of specific organs and tissues. It is subsequently silenced in most adult tissues but can be reactivated following injury to promote repair and regeneration.
Different Hh ligands, including homologs sonic hedgehog (SHH), indian hedgehog (IHH), and desert hedgehog (DHH) are produced as precursors that undergo autocatalytic cleavage, C-terminal cholesterol attachment, and N-terminal palmitoylation prior to secretion. Release and extracellular accumulation of the mature ligands is regulated by the homologs of the Drosophila dispatched (Disp) protein. Binding of secreted Hh ligands to the receptor homolog (PTCH1, PTCH2) triggers the stimulation of the signaling network: the repressive effect of PTCH on the transmembrane receptor SMO is relieved which leads to the activation of glioma-associated oncogene (GLI) transcription factors.
A key specialized structure in this process is the microtubule-based primary cilium. In the absence of the Hh signal unprocessed, non-activating GLI proteins as well as their regulator SUFU are concentrated in the distal tip of the primary cilium. Upon binding of the Hh ligand, PTCH relocates to the cell surface, thus rendering translocation of SMO to the primary cilium and the down-regulation of SUFU possible. Several structurally essential primary cilium proteins have also regulatory effects on the Hh signaling cascade.
Aberrant Hh/GLI regulation leads to major tissular disorders and the development of a wide variety of aggressive cancers. The Hh/GLI cascade has also been linked to the regulation of stemness genes and the survival of cancer stem cells.
References
- : "Hedgehog Pathway Inhibitors: A New Therapeutic Class for the Treatment of Acute Myeloid Leukemia." in: Blood cancer discovery, Vol. 1, Issue 2, pp. 134-145, (2022) (PubMed).
- : "The emerging conceptual framework of evolutionary developmental biology." in: Nature, Vol. 415, Issue 6873, pp. 757-64, (2002) (PubMed).
- : "Posttranscriptional regulation of smoothened is part of a self-correcting mechanism in the Hedgehog signaling system." in: Molecular cell, Vol. 6, Issue 2, pp. 457-65, (2000) (PubMed).
- : "Anti-patched-1 antibodies suppress hedgehog signaling pathway and pancreatic cancer proliferation." in: Anticancer research, Vol. 27, Issue 6A, pp. 3743-7, (2007) (PubMed).
- : "The hedgehog signaling pathway: where did it come from?" in: PLoS biology, Vol. 7, Issue 6, pp. e1000146, (2009) (PubMed).
- : "The Hedgehog protein family." in: Genome biology, Vol. 9, Issue 11, pp. 241, (2009) (PubMed).
- : "Transducing the hedgehog signal." in: Cell, Vol. 103, Issue 3, pp. 371-4, (2000) (PubMed).
- : "The hedgehog signaling network." in: American journal of medical genetics. Part A, Vol. 123A, Issue 1, pp. 5-28, (2004) (PubMed).
- : "Hedgehog/Gli control by ubiquitination/acetylation interplay." in: Vitamins and hormones, Vol. 88, pp. 211-27, (2012) (PubMed).