Epigenetics is the study of heritable cellular, and physiological traits by factors other than DNA sequence. Examples are simple chemical modifications, like methylation and acetylation to DNA and interacting partners. These changes are reversible and do not directly change the nucleotide sequence. However they play a role in controlling how, where, and when certain genes are expressed.
These epigenetic changes may last through cell divisions for the duration of the cell's life, and may also last for multiple generations even though they do not involve changes in the underlying DNA sequence of the organism. Epigenetic research has the potential to explain mechanisms of aging, human development, and the origins of several health conditions, e.g. cancer, heart disease and mental illness.
Mapping Protein-DNA Interactions with ChIP
Immunoprecipitation of protein-DNA complexes (ChIP) enables the study of epigenetic relations. The technique identifies binding sites of DNA-associated proteins and allows a closer study of transcription factors. Targeted nuclease cleavage and release is followed by chromatin immunoprecipitation. The ChIP uses antibodies that selectively recognize, bind and isolate proteins, including histones, histones with modifications, transcription factors and cofactors, similar to conventional IP. Linked gene sequences may then be identified via high throughput DNA sequencing which is referred to as ChIP-Seq.
Variations of ChIP
Several variations are established for ChIP. In X-ChIP, formaldehyde cross-links the protein of interest with DNA. Fragmentation of the chromatin is achieved by ultrasound treatment or nuclease digestion.
For Native ChIP, chromatin is isolated from cell nuclei that have been digested with a nuclease. Then the chromatin is sheared by micrococcal nuclease digestion, which cuts DNA at the length of the linker.
Native ChIP
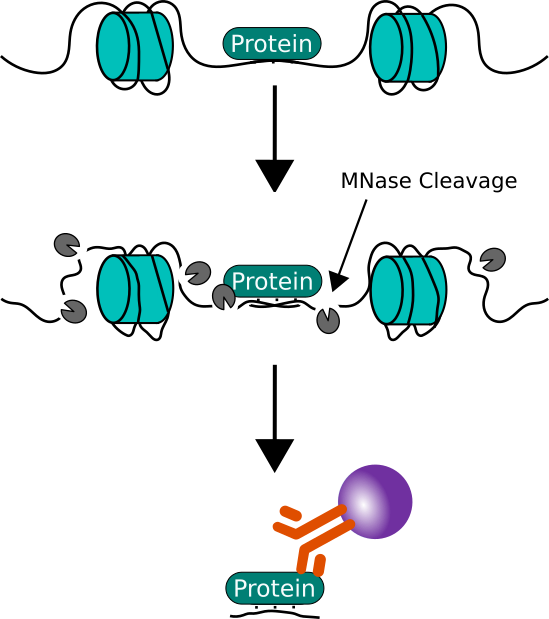
Advantages:
- Testable Antibody Specificity
- Better Antibody specificity through native Protein
- Better Chromatin and Protein recovery
Disadvantages:
- Not suitable for non-histone Proteins
- Nucleosomes may rearrange during digestion
Cross-linked ChIP

Advantages:
- Suitable for Transcriptional Factors
- Applicable to any Organism
Disadvantages:
- Inefficient Chromatin Recovery
- May cause false positive Result
- Random Cut by Sonication leads to wide Variance in Chromatin Length
CUT&RUN - Superior Alternative to ChIP-Seq
CUT&RUN-sequencing, also known as Cleavage Under Targets and Release Using Nuclease, is a method used to analyze protein interactions with DNA. CUT&RUN-sequencing combines antibody-targeted controlled cleavage by micrococcal nuclease with massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins.
Advantages of CUT&RUN
- No Crosslinking, the factor-bound DNA that is cleaved on both sides of the bound particle is efficiently released into solution
- Limited digestion of chromatin only aroundtarget sites minimizes fragments
- Better signal to background noise ratio
- greatly reducing sequencing depth
- Application viable with limited sample amount
Need Help? Call our PhD Customer Support!
- We help you with finding the right product for your research.
- We offer reliable antibodies, kits, proteins, lysates for research.
- Contact us via email or phone: (877) 302 8632 (US) or +49 241 95 163 153 (International)
Popular Antibodies for ChIP
- (1)
- (9)
- (1)
- (7)
- (20)
- (1)
- (8)
- (14)
- (1)
- (1)
- (4)
- (1)
- (4)
- (21)
- (1)
- (23)
- (5)
- (1)
- (4)
- (1)
- (3)
- (5)
- (1)
- (4)
- (5)
- (1)
- (14)
- (17)
- (75)
- (5)
- (6)
- (8)
- (2)
- (8)
- (5)
- (15)
- (7)
- (17)

Creative mind of antibodies-online with a keen eye for details. Proficient in the field of life-science with a passion for plant biotechnology and clinical study design. Responsible for illustrated and written content at antibodies-online as well as supervision of the antibodies-online scholarship program.
Go to author page



